Title: The CLASSY family controls tissue-specific DNA methylation patterns in Arabidopsis
Ming Zhou, Ceyda Coruh, Guanghui Xu, Laura M. Martins, Clara Bourbousse, Alice Lambolez & Julie A. Law
Nature Communications volume 13, Article number: 244 (2022) Cite this article
Abstract
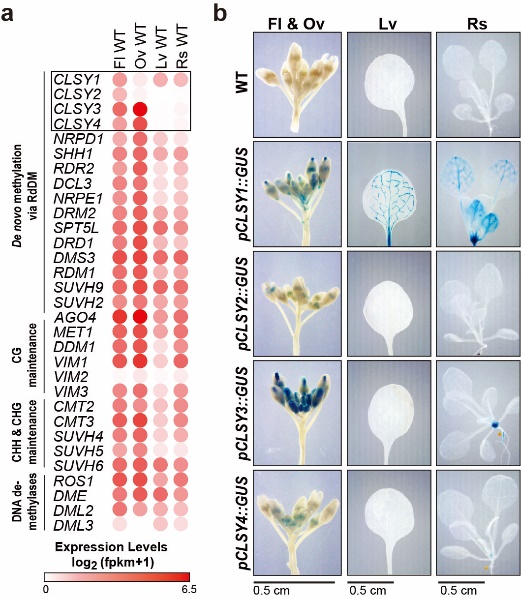
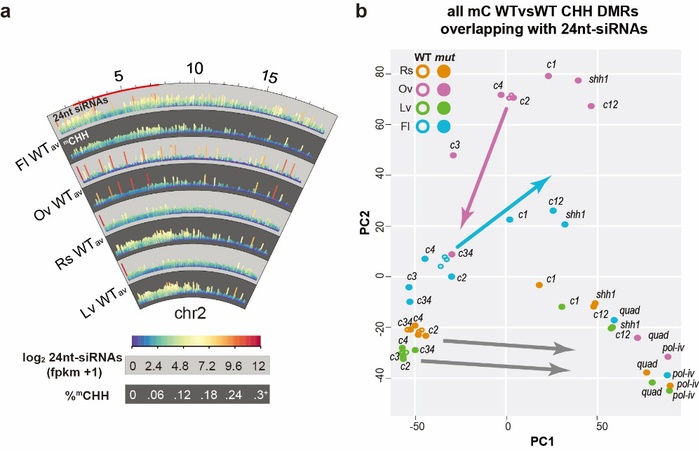
DNA methylation shapes the epigenetic landscape of the genome, plays critical roles in regulating gene expression, and ensures transposon silencing. As is evidenced by the numerous defects associated with aberrant DNA methylation landscapes, establishing proper tissue-specific methylation patterns is critical. Yet, how such differences arise remains a largely open question in both plants and animals. Here we demonstrate that CLASSY1-4 (CLSY1-4), four locus-specific regulators of DNA methylation, also control tissue-specific methylation patterns, with the most striking pattern observed in ovules where CLSY3 and CLSY4 control DNA methylation at loci with a highly conserved DNA motif. On a more global scale, we demonstrate that specific clsy mutants are sufficient to shift the epigenetic landscape between tissues. Together, these findings reveal substantial epigenetic diversity between tissues and assign these changes to specific CLSY proteins, elucidating how locus-specific targeting combined with tissue-specific expression enables the CLSYs to generate epigenetic diversity during plant development.
Link: https://www.nature.com/articles/s41467-021-27690-x