PNAS
Title: Global genomic instability caused by reduced expression of DNA polymerase ε in yeast
Ke Zhang, Yang Sui, Wu-Long Li, Gen Chen, Xue-Chang Wu, Robert J. Kokoska, Thomas D. Petes tom.petes, and Dao-Qiong Zheng
March 15, 2022 | 119 (12) e2119588119 | https://doi.org/10.1073/pnas.2119588119
Abstract
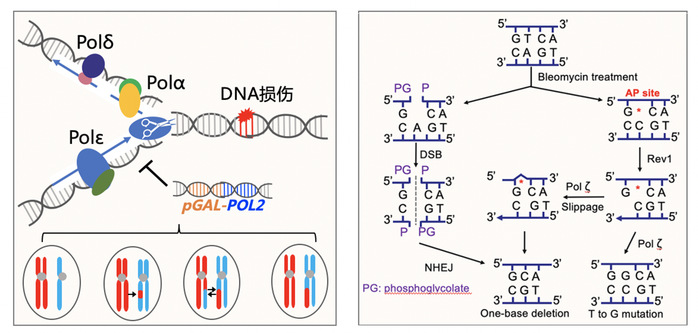
DNA polymerase ε (Pol ε) is one of the three replicative eukaryotic DNA polymerases. Pol ε deficiency leads to genomic instability and multiple human diseases. Here, we explored global genomic alterations in yeast strains with reduced expression of POL2, the gene that encodes the catalytic subunit of Pol ε. Using whole-genome SNP microarray and sequencing, we found that low levels of Pol ε elevated the rates of mitotic recombination and chromosomal aneuploidy by two orders of magnitude. Strikingly, low levels of Pol ε resulted in a contraction of the number of repeats in the ribosomal DNA cluster and reduced the length of telomeres. These strains also had an elevated frequency of break-induced replication, resulting in terminal loss of heterozygosity. In addition, low levels of Pol ε increased the rate of single-base mutations by 13-fold by a Pol ζ-dependent pathway. Finally, the patterns of genomic alterations caused by low levels of Pol ε were different from those observed in strains with low levels of the other replicative DNA polymerases, Pol α and Pol δ, providing further insights into the different roles of the B-family DNA polymerases in maintaining genomic stability.
Link: https://www.pnas.org/doi/abs/10.1073/pnas.2119588119

AEM
Title: Uncovering Bleomycin-Induced Genomic Alterations and Underlying Mechanisms in the Yeast Saccharomyces cerevisiae
Dao-Qiong Zheng, Yu-Ting Wang, Ying-Xuan Zhu, Huan Sheng, Ke-Jing Li, Yang Sui, Ke Zhang
DOI: https://doi.org/10.1128/AEM.01703-21
Abstract
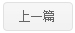
Bleomycin (BLM) is a widely used chemotherapeutic drug. BLM-treated cells showed an elevated rate of mutations, but the underlying mechanisms remained unclear. In this study, the global genomic alterations in BLM-treated cells were explored in the yeast Saccharomyces cerevisiae. Using genetic assay and whole-genome sequencing, we found that the mutation rate could be greatly elevated in S. cerevisiae cells that underwent Zeocin (a BLM member) treatment. One-base deletion and T-to-G substitution at the 5′-GT-3′ motif represented the most striking signature of Zeocin-induced mutations. This was mainly the result of translesion DNA synthesis involving Rev1 and polymerase ζ. Zeocin treatment led to the frequent loss of heterozygosity and chromosomal rearrangements in the diploid strains. The breakpoints of recombination events were significantly associated with certain chromosomal elements. Lastly, we identified multiple genomic alterations that contributed to BLM resistance in the Zeocin-treated mutants. Overall, this study provides new insights into the genotoxicity and evolutional effects of BLM.
Link: https://journals.asm.org/doi/full/10.1128/AEM.01703-21