Title: Trans-splicing facilitated by RNA pairing greatly expands sDscam isoform diversity but not homophilic binding specificity
SHOUQING HOU, GUO LI, BINGBING XU, HAIYANG DONG, SHIXIN ZHANG, YING FU, JILONG SHI, LEI LI, JIAYAN FU, FENG SHI, YIJUN MENG AND YONGFENG JIN
Abstract
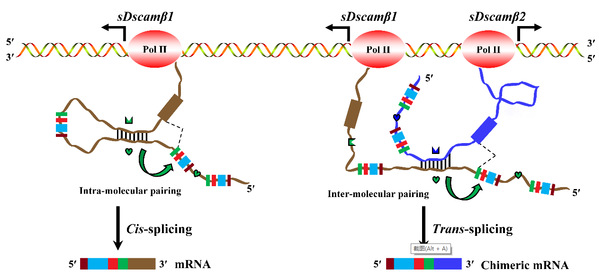
The Down syndrome cell adhesion molecule 1 (Dscam1) gene can generate tens of thousands of isoforms via alternative splicing, which is essential for nervous and immune functions. Chelicerates generate approximately 50 to 100 shortened Dscam (sDscam) isoforms by alternative promoters, similar to mammalian protocadherins. Here, we reveal that trans-splicing markedly increases the repository of sDscamβ isoforms in Tetranychus urticae. Unexpectedly, every variable exon cassette engages in trans-splicing with constant exons from another cluster. Moreover, we provide evidence that competing RNA pairing not only governs alternative cis-splicing but also facilitates trans-splicing. Trans-spliced sDscam isoforms mediate cell adhesion ability but exhibit the same homophilic binding specificity as their cis-spliced counterparts. Thus, we reveal a single sDscam locus that generates diverse adhesion molecules through cis- and trans-splicing coupled with alternative promoters. These findings expand understanding of the mechanism underlying molecular diversity and have implications for the molecular control of neuronal and/or immune specificity.
Link: https://www.science.org/doi/10.1126/sciadv.abn9458