Title: Unfolding of RNA secondary structure impairs RNA stability to fine-tune phosphate starvation responses in rice roots
Qiongli Jin, Ruiren Gao, Jiakai Yao, Zhengwei Huang, Kai Liu, Guangbo Wei, Weiguo Dong, Zhiye Wang
Abstract
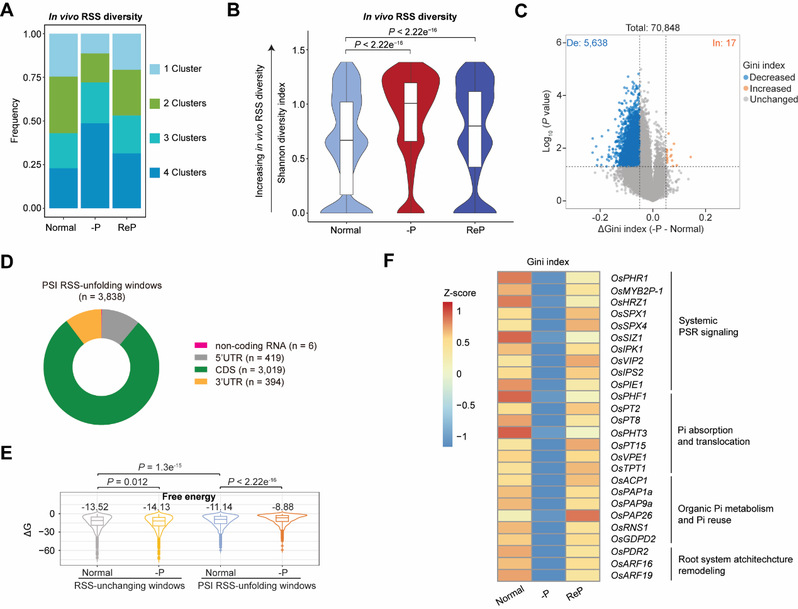
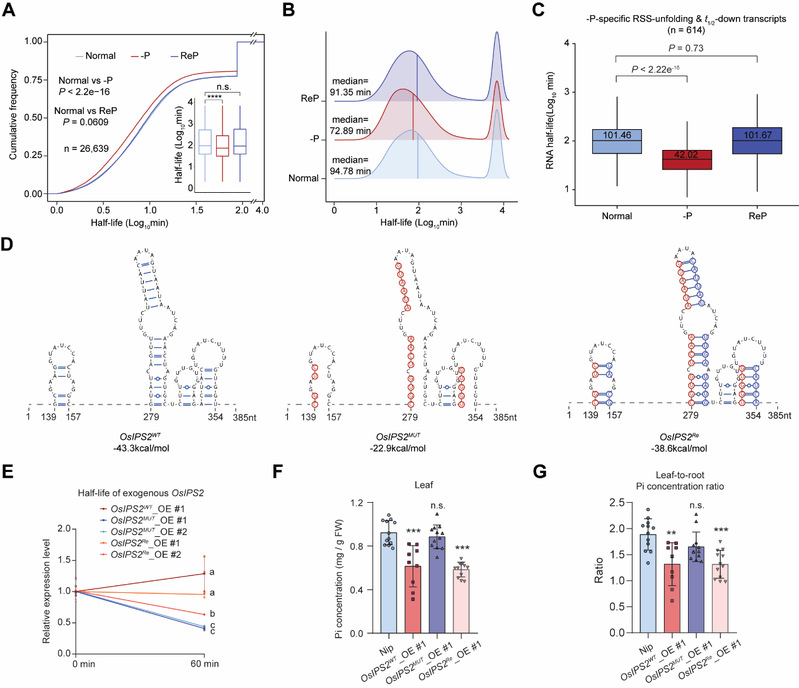
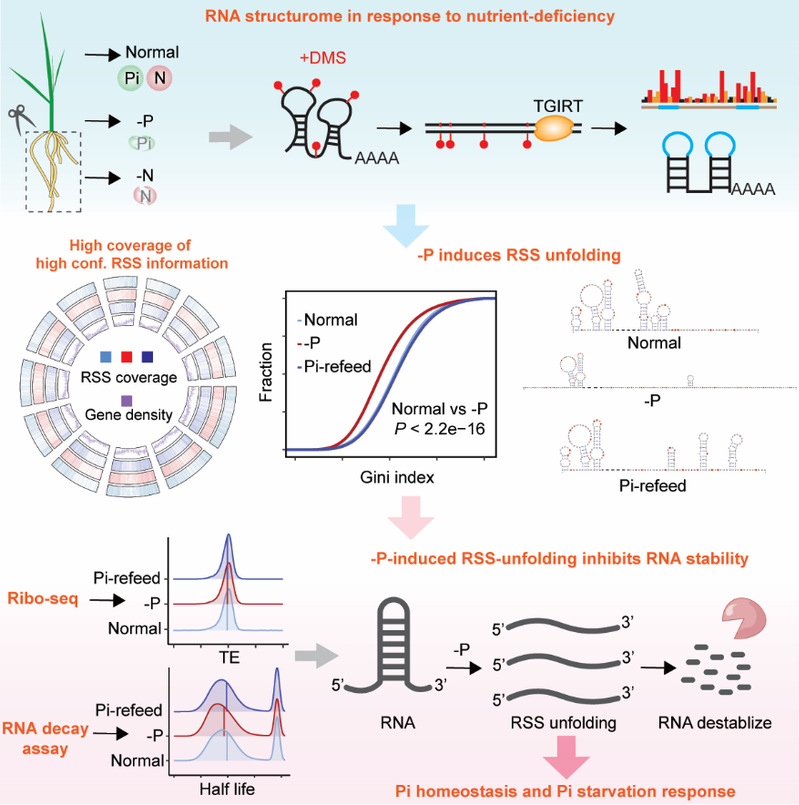
The availability of essential macronutrients such as inorganic phosphate (Pi) and nitrogen limits global crop production. In vivo RNA secondary structure (RSS) regulates almost all steps in the RNA life cycle and is dynamic under stress conditions. However, the effects of RSS in plants in response to nutrient deficiency stress remain unclear. Here, we used dimethyl sulfate mutational profiling with sequencing (DMS-MaPseq) to generate high-quality, deep-coverage in vivo RSS profiles of rice (Oryza sativa) roots in response to Pi deficiency (–P) or nitrogen deficiency (–N) stress. –P increased global RSS diversity and triggered RSS unfolding in thousands of transcripts. By comparing the –P and –N RSS profiles, we identified –P-specific RSS-unfolding regions in rice roots. These regions, which had low GC content, were present in the CDS of many Pi starvation response transcripts. Ribosome profiling suggested that –P-specific RSS unfolding was not linked to translational regulation. By contrast, transcriptome-wide RNA decay assays under normal, –P, and Pi-refeeding conditions revealed global regulation of RNA stability in response to –P in rice roots and decreases in RNA half-life with RSS unfolding. Transcriptome analysis and analysis of transgenic rice plants altered in RSS revealed that –P-specific RSS unfolding lowers RNA stability, thereby fine-tuning Pi-starvation response transcripts’ accumulation and the Pi homeostasis. This study systemically elucidates the dynamic roles and the regulatory functions of RSS in the Pi-starvation response in rice roots. These findings underscore the important role of RSS in modulating nutrition-deficient stress responses.
Link: https://www.cell.com/plant-communications/fulltext/S2590-3462(25)00442-0